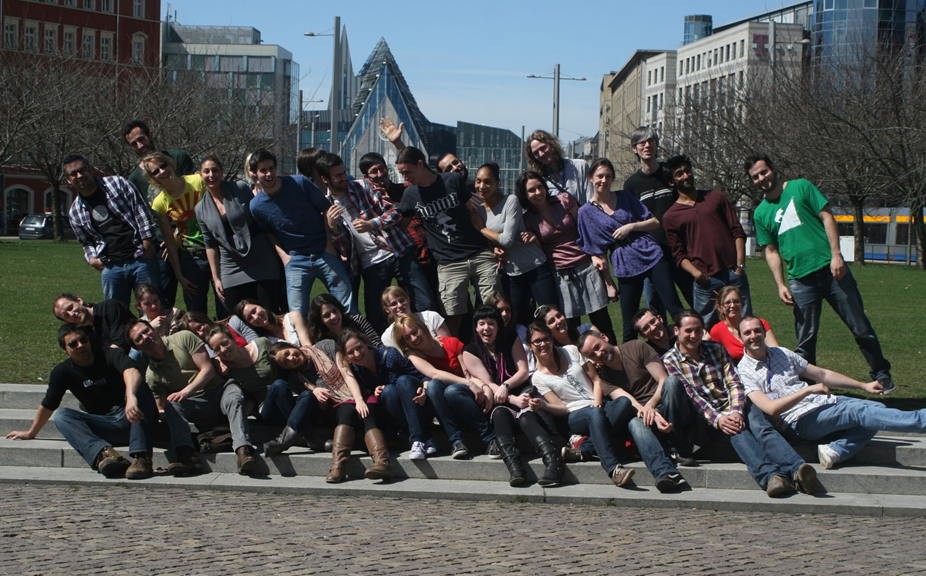3rd Programming for Evolutionary Biology Course
March 21st – April 6th 2014 Leipzig, Germany
Modules and Instructors
Introduction to Linux systems
Giovanni Marco Dall’Olio, University Pompeu Fabra, Barcelona, Spain
Analysis of high-throughput sequencing data
David Langenberger, Steve Hoffmann, University of Leipzig, Germany
Analysis of RAD sequencing data and Assembly
Anja Westram, University Sheffield, United Kingdom
Introduction to R
Katja Nowick, University of Leipzig, Germany
Comparative analysis of expression data
Mehmet Somel, METU Department of Biological Sciences, Ankara, Turkey
Population genomics
Johannes Engelken, University Pompeu Fabra, Barcelona, Spain
Statistics & Inference
Stuart Baird, University of Antwerp, Belgium
Introduction to Perl
Sofia Robb, University of California Riverside, USA
Introduction to BioPerl
Sofia Robb, University of California Riverside, USA
Phylogenomics
Christoph Bleidorn, University of Leipzig, Germany
Introduction to databases
Jan Aerts, Leuven University, Belgium
Ensembl API
Bert Overduin, EMBL – European Bioinformatics Institute, Hinxton, Cambridge, UK
Visualization of scientific data
Jan Aerts, Leuven University, Belgium
3 days final project
Invited speakers
Structural variants in primates
Tomas Marques-Bonet, University Pompeu Fabra, Barcelona, Spain
The genomics of adaptive radiation
Walter Salzburger, University of Basel, Switzerland
Genomics of speciation
Rui Faria, University of Porto, Portugal
Epistasis and molecular evolution
Fyodor Kondrashov, University Pompeu Fabra, Barcelona, Spain
Evolution of non-coding RNAs
Peter Stadler, University of Leipzig, Germany
Teaching Assistants
Lydia Steiner, University of Leipzig, Germany
Jan Engelhardt, University of Leipzig, Germany
Alvaro Perdomo-Sabogal, University of Leipzig, Germany
Sree Rohit Raj Kolora, University of Leipzig, Germany
Tiago Carvalho, University Pompeu Fabra, Barcelona, Spain
Vladimir Jovanovic, University of Belgrade, Serbia
Rik Verdonck, University of Leuven, Belgium
Technical Support
Jens Steuck, University of Leipzig, Germany
Valentina Klebanova, DITON, Regis-Breitingen, Germany
2nd Course: April 3rd – April 19th 2013
Modules and Instructors
Introduction to Linux systems
Giovanni Marco Dall’Olio, University Pompeu Fabra, Barcelona, Spain
Analysis of high-throughput sequencing data (with reference genome)
Steve Hoffmann, David Langenberger, University of Leipzig, Germany
Analysis of high-throughput sequencing data (for non-model organisms)
Sonja Grath, University of Munich (LMU), Germany
Introduction to R
Katja Nowick, University of Leipzig, Germany
Comparative analysis of expression data
Mehmet Somel, University of California, Berkeley, USA
Biological graphs and networks
Jing Qin, University of Leipzig, Germany
Introduction to Perl
Sofia Robb, University of California Riverside, USA
Introduction to BioPerl
Sofia Robb, University of California Riverside, USA
Phylogenomics
Christoph Bleidorn, University of Leipzig, Germany
Population genomics
Johannes Engelken, University Pompeu Fabra, Barcelona, Spain
Introduction to databases
Jan Aerts, Leuven University, Belgium
Visualization of scientific data
Jan Aerts, Leuven University, Belgium
4 days final project
Invited speakers
Evolution of behavior
Sarah London, University of Chicago, USA
Analysis of structural variants
Tomas Marques-Bonet, University Pompeu Fabra, Barcelona, Spain
Genomics of speciation
Rui Faria, University of Porto, Portugal
Evolution of non-coding RNAs
Peter Stadler, University of Leipzig, Germany
Teaching Assistants
Lydia Steiner, University of Leipzig, Germany
Jan Engelhardt, University of Leipzig, Germany
Miguel Fonseca, University of Vigo, Spain
Tiago Carvalho, University Pompeu Fabra, Barcelona, Spain
Vladimir Jovanovic, University of Belgrade, Serbia
Rik Verdonck, University of Leuven, Belgium
Technical Support
Jens Steuck, University of Leipzig, Germany
Petra Pregel, University of Leipzig, Germany
Valentina Klebanova, DITON, Regis-Breitingen, Germany
1st Course: March 17th – April 1st 2012
Modules and Instructors
Introduction to Linux systems
Giovanni Marco Dall’Olio, University Pompeu Fabra, Barcelona, Spain
Introduction to R
Katja Nowick, University Leipzig, Germany
Analysis of next generation sequencing data
David Langenberger, University Leipzig, Germany
Analysis of structural variants
Tomas Marques-Bonet, University Pompeu Fabra, Barcelona, Spain
Analysis of expression data
Katja Nowick, University Leipzig, Germany
Promoter evolution
Annalisa Marsico, Max-Planck-Institute for Molecular Genetics, Berlin, Germany
Statistics & Inference
Stuart Baird, University Porto, Portugal
Introduction to Perl
Sofia Robb, University of California Riverside, USA
Phylogenomics
Rasmus Nielsen, University of California, Berkeley, USA
Rui Faria, University Porto, Portugal
Ensembl API
Bert Overduin, EMBL – European Bioinformatics Institute, Hinxton, Cambridge, UK
Introduction to databases
Jan Aerts, Leuven University, Belgium
Visualization of scientific data
Jan Aerts, Leuven University, Belgium
3 days final project
Invited speakers
Evolution of behavior
Sarah London, University of Chicago, USA
Evolutionary ecology
Claudia Acquisti, Westfälische Wilhelms-University Münster, Germany
Genotype to phenotype mapping
Ben Lehner, Centre for Genomic Regulation, Barcelona, Spain
NGS of non-model organisms
Markus Pfenninger, Biodiversity and Climate Research Center, Frankfurt, Germany
Teaching Assistants
Lydia Steiner – University Leipzig, Germany
Christian Arnold – University Leipzig, Germany
Rosalina Tincopa – University Leipzig, Germany
Stefano Berto – University Leipzig, Germany
Tiago Carvalho – University Porto, Portugal
Miguel Fonseca – University Porto, Portugal
Javier Prado – University Pompeu Fabra, Barcelona, Spain
Technical Support
Jens Steuck
Petra Pregel
See more pictures on our Facebook page!